NNucleate’s documentation
NNucleate is a Python package developed for the training of approximations of collective variables (CV). The primary intended application is enhanced sampling simulations of nucleation events. These types of simulations are typically limited by the computational cost of their CVs, since they require a computationally expensive, differentiable degree of order in the system to be calculated on the fly. When using this code please cite:
10.26434/chemrxiv-2023-l6jjd
Installation
The package can be installed using pip:
pip install git+https://github.com/Flofega/NNucleate.git
Overview
Approximations for collective variables that need to be permutationally invariant can be built using the class NNucleate.models.GNNCV. These approximations are based on a graph neural network architecture which is inherently permutationally invariant.
The main cost advantage compared to the approximated variables is gained by mapping directly from cartesian coordinates to local contributions to the global CV value, skipping the need for expensive descriptors or symmetry functions.
For a more detailed discussion please refer to the paper.
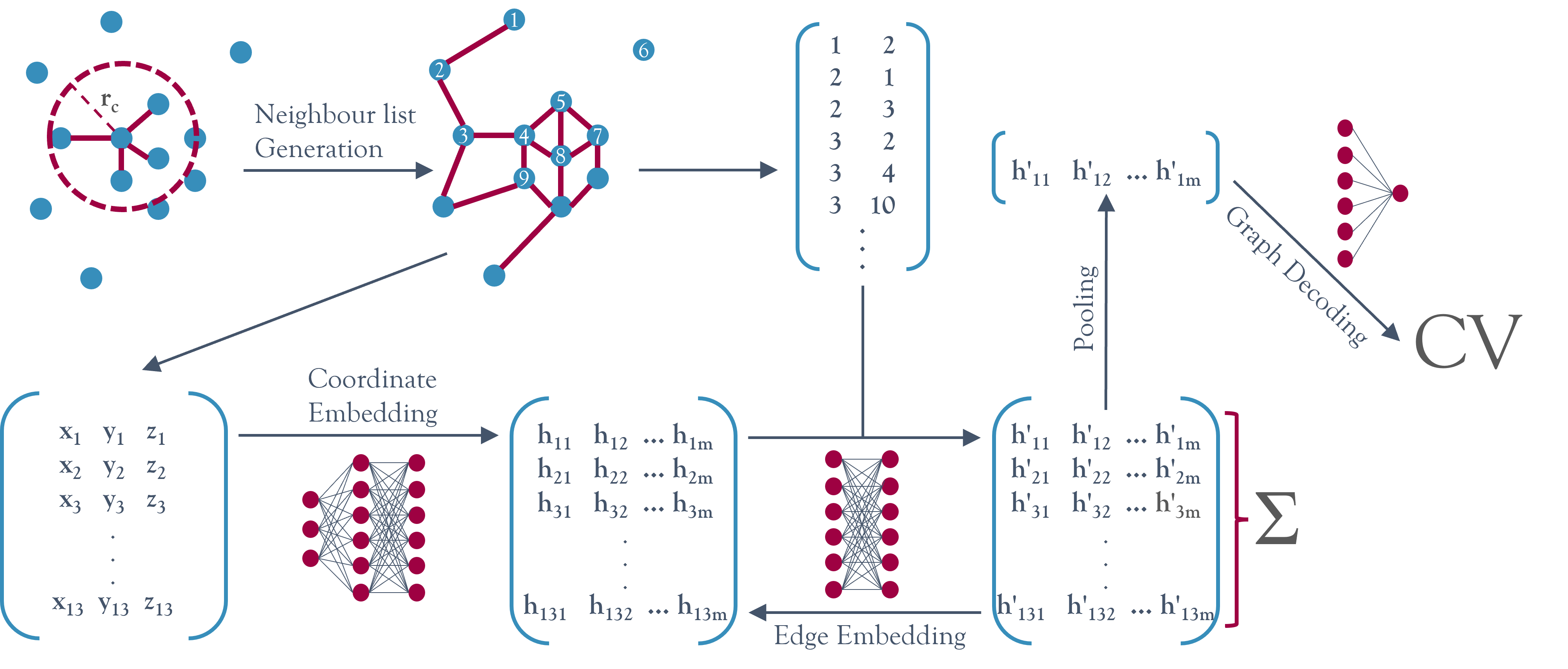
Approximations that do not have to be permutationally invariant (e.g. when working with single proteins), can be constructed using the class NNucleate.models.NNCV. These types of approximations based on linear neural networks are an order of magnitude faster than the GNN based ones but require more training data, do not generalise outside of their training domain and are not size-transferable.
Basic Tutorial
This package is built on top of PyTorch and the user experience is designed to deviate as little as possible from a typical Pytorch project.
Start by loading the training data into your project using the appropriate dataset class from NNucleate.dataset and creating a wrapper object around the dataset:
from NNucleate.dataset import GNNTrajectory
from torch.utils.data import Dataset, DataLoader
ds = GNNTrajectory(cv_file_path, trajectory_data_path, topology_file_path, cv_file_column, box_length, cut_off, stride=1, root=1)
dataloader = DataLoader(ds, batch_size=64, shuffle=True)
Then initialise the model, optimiser and loss function of choice and use the functionalities from NNucleate.training to train the model until convergence:
from NNucleate.models import GNNCV
from NNucleate.training import train_gnn, test_gnn
from torch import optim, nn
model = GNNCV(hidden_nf=25, n_layers=3)
optimizer = optim.Adam(model.parameters(), lr=5e-4)
loss = nn.MSELoss()
for epoch in range(epochs):
train_mse = train_gnn(model, dataloader, n_at, optimizer, loss, device))
if epoch % 5 == 0:
test_mse = test_gnn(model, test_dataloader, n_at, loss, device))
print("Epoch %d" % epoch)
print("Training Error: %f" % train_mse)
print("Test set Error: %f" % test_mse)
Some basic metrics and visualisations for the performance of the final model can be obtained by calling NNucleate.training.evaluate_model_gnn():
import matplotlib.pyplot as plt
from NNucleate.training import evaluate_model_gnn
preds, labels, rmse, r2 = evaluate_model_gnn(model, validation_dataloader, n_at, device)
plt.scatter(preds, labels, s=0.4)
plt.title("Train: r = %.4f" % r2)
plt.xlabel("Prediction")
plt.ylabel("Label")
plt.show()
The graph-based approximations can be deployed as collective variables using the Plumed fork PyCV. The helper function NNucleate.pycv_link.write_cv_link() can turn the model object into a PyCV input file:
from NNucleate.pycv_link import write_cv_link
write_cv_link(model, hidden_nf, n_layers, n_at, box_l, rc, "gnn_cv.py")
This file contains a function “cv1” which can be read and used by Plumed as follows:
gnnCV: PYTHONCV ATOMS=1-500 IMPORT=gnn_cv FUNCTION=cv1
Approximations based on linear neural networks can be implemented directly using the “annfunc” module in Plumed.
The whole training process can be done on a GPU by specifying the device variable and moving the necessary objects and arrays to the device using .to(device).